On July 30, 2022, Prof. Fan Xiaohui's team at the Future Health Laboratory of the Yangtze River Delta Innovation Center of Zhejiang University, in collaboration with Prof. Xu Xiao's team at the First People's Hospital of Hangzhou affiliated to the Medical College of Zhejiang University, and Prof. Chen Huajun's team at the School of Computer Science of Zhejiang University, published an online research paper in the journal Nature Communications Knowledge-graph-based cell-cell communication inference for spatially resolved transcriptomic data with SpaTalk.

Intercellular communication, also known as intercellular interaction, refers to the process by which one cell generates a signal that is transmitted to another cell through a medium, which in turn regulates the target cell and produces a series of biological effects such as growth, proliferation, differentiation, and apoptosis. Accurate identification of intercellular communication is a key link in understanding disease development and developing interventional drugs, which is increasingly emphasized by the industry.
With the development of recent years, the rapid development of spatially resolved transcriptomics sequencing technology has been widely used to explore the spatial heterogeneity of cells, as it realizes both transcriptome sequencing and preserves the spatial location information of cells and their genes. Currently, spatial transcriptome sequencing technologies are divided into two main categories according to their principles. The first category is based on high-throughput RNA imaging technologies, including STARmap, MERFISH, seqFISH, etc., which realize spatial transcriptome sequencing at single-cell resolution; the second category is based on spatial barcoding technologies, including 10X Visium, Slide-seq, HDST, Stereo-seq, etc., which realize spatial regions of whole transcriptome sequencing. . Obviously, the rapid development of spatial transcriptome sequencing technology provides technical support and data sources for studying the spatial heterogeneity of intercellular communication, and how to utilize spatial transcriptome data to infer intercellular communication has become an important challenge in related fields.
In this study, we developed SpaTalk, a spatial transcriptome sequencing data-based method for inferring intercellular communication, by introducing artificial intelligence algorithms such as knowledge graph, non-negative linear regression, and random walk, etc. The method integrates the spatial proximity principle of ligand-receptor and co-expression of ligand-receptor and downstream targets, and performs graph network and knowledge graph modeling and scoring on spatially significant ligand-receptor and signaling pathways in the receptor cells, respectively. knowledge graph modeling and scoring to infer ligand-receptor interaction-mediated intercellular communication in a restricted space.
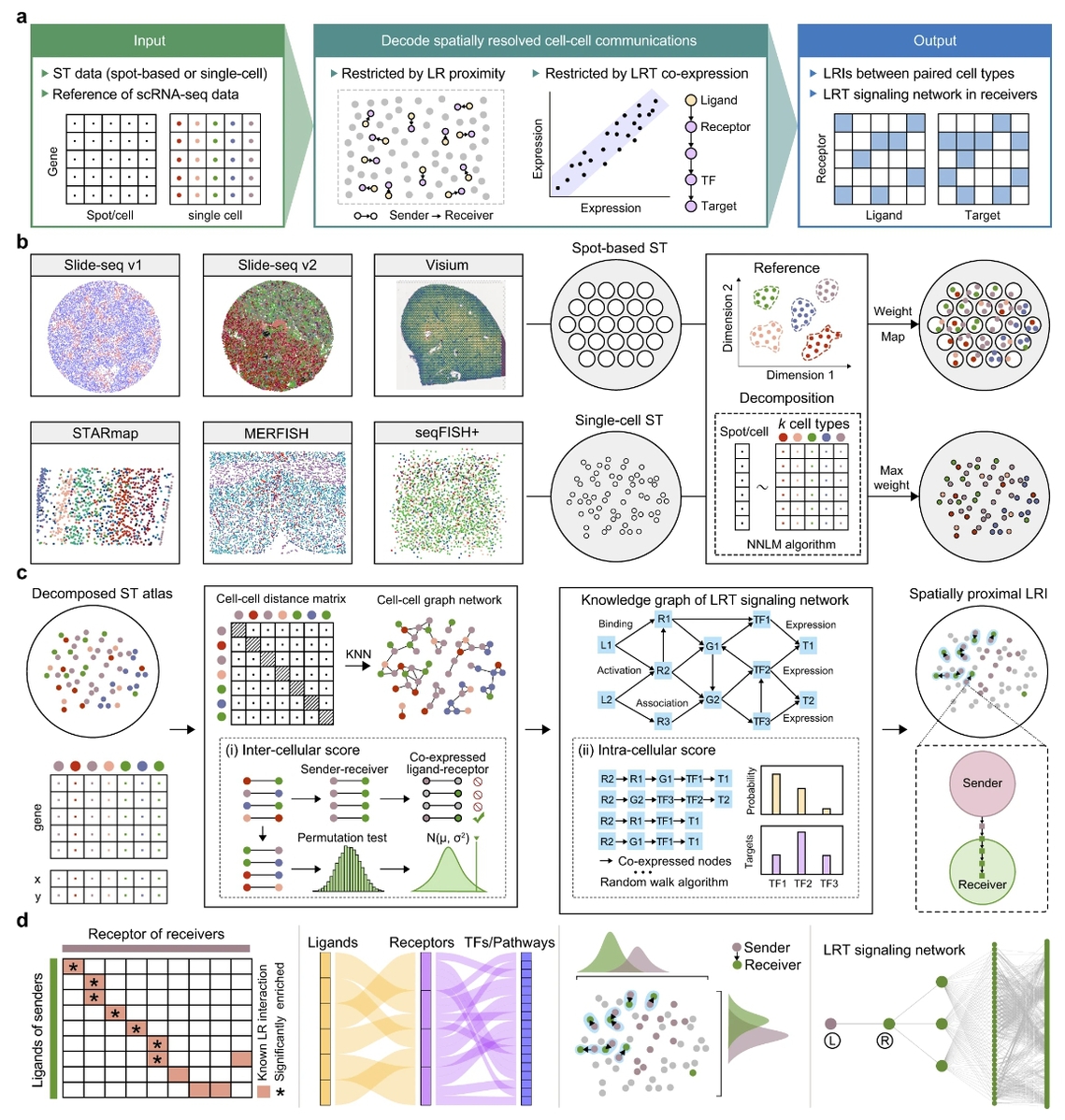
SpaTalk is divided into two steps: (1) The first step is to use the single-cell transcriptome sequencing data as a reference map to resolve the cellular composition in the spatial transcriptome data, and then reconstruct the single-cell map with known cell types and spatial coordinates. For single-cell resolution idling data, SpaTalk identifies the type of each single cell in the idling data through non-negative linear regression; for regional resolution idling data, SpaTalk identifies the proportion of different cell types in each region through non-negative linear regression, and thus utilizes spatial mapping to reduce the single cells in the reference atlas to different spatial regions. (2) The second step is to model ligand-receptor-downstream transcription factor-target genes using knowledge mapping, and filter ligand-receptor interaction pairs that are spatially significantly adjacent and activated by downstream signals of the receptor cells through algorithms such as substitution test and random walk.
The results show that SpaTalk can not only accurately resolve the cell proportions of the baseline test data, which is better than other deconvolution methods; it can also accurately infer the ligand-receptor pairs that mediate spatial cell-to-cell communication and their activated downstream signaling pathways, which is more accurate than other methods for inferring intercellular communication. Compared with other existing methods, SpaTalk enables the comparison and visualization of intercellular ligand-receptor interactions at the single-cell level for the first time, and the application cases further demonstrate the unique advantages of SpaTalk in resolving the key intercellular communications in normal physiological and pathological processes at the spatial single-cell resolution.
This research was funded by the National Natural Science Foundation of China, the National Key Research and Development Program of China, and the Natural Science Foundation of Zhejiang Province, and supported by Aliyun.


